
Some call it a swarm of variants — others refer to it as variant soup. Whatever it’s called, the current crop of immunity-dodging offshoots of the Omicron variant of SARS-CoV-2 is unprecedented in its diversity. This complexity makes it harder to predict coming waves of infection. It might even lead to a ‘double wave’ in some places, as first one variant and then another overtakes a population.
But amid the chaos, patterns are emerging. The swarm has helped scientists to pinpoint a handful of immunity-evading mutations that power a variant’s spread. Globally, a few heavyweight variants have emerged, yielding different outcomes in different regions — at least, so far.
In Europe, North America and Africa, the prevalence of Omicron offshoots in the BQ.1 family is rising quickly, even as overall cases seem to fall. In Asian countries including Singapore, Bangladesh and India, a lineage called XBB has already set off fresh waves of infection. Scientists are closely watching several regions where both are circulating, to see which has the edge.
“In the end, probably, some variants are going to dominate, but it’s less decisive than it was in the past,” says Cornelius Roemer, a computational biologist at the University of Basel in Switzerland.
One big family
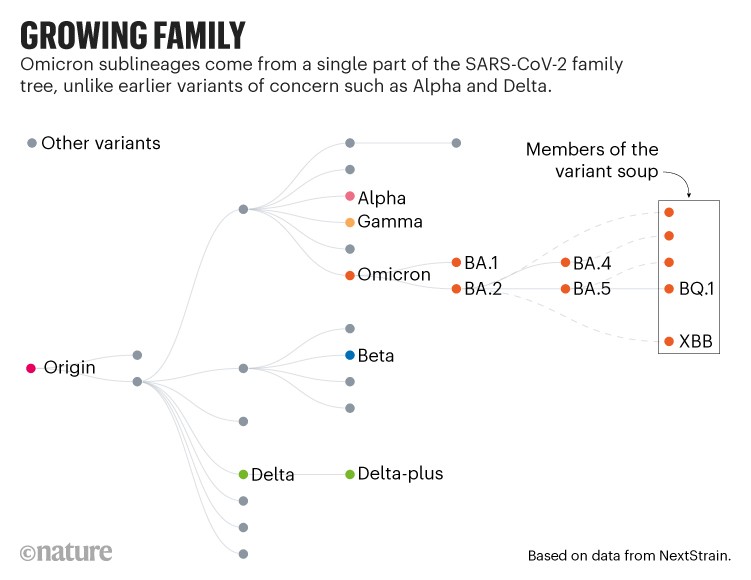
The variants that have driven past waves, such as Alpha and Delta, all arose from distinct branches of the SARS-CoV-2 family tree. But since Omicron emerged in late 2021, it has spawned a series of subvariants, including BA.2 and BA.5, that have sparked global waves of infection. Many countries put their BA.5-led surges in the rear-view mirror in mid-2022, but most scientists thought it was only a matter of time before another sublineage came to the fore.
For the past few months, variant trackers have been combing through global SARS-CoV-2 sequencing data to identify candidates. But instead of one or two fast-rising lineages, they have identified more than a dozen to watch.
“It’s a collection or swarm or soup of variants together — not as we have seen before,” says Yunlong Richard Cao, an immunologist at Peking University in Beijing, whose team has been studying the variants’ immune-evading capacities.
The members of the swarm come from various parts of the Omicron family tree. But their rise seems to be due to a handful of shared genetic mutations, most of which lead to amino acid changes in a portion of the viral spike protein called the receptor binding domain (RBD). This part of the protein is required to infect cells, and is the target of antibodies that deliver a potent immune response.
Work from Cao’s team this month1 suggests that the RBD mutations help the virus to evade infection-blocking ‘neutralizing’ antibodies that were triggered by COVID-19 vaccines and infection with earlier Omicron offshoots, including BA.2 and BA.5. (That work has not yet been peer reviewed.)
Ringing the changes
Roemer and others have observed that the more of these RBD changes a variant possesses, the faster it seems to grow, as measured by the number of sequences reported to global databases. For instance, variants, such as BQ.1,with five key RBD changes (relative to BA.2) seem to be growing in number at a slower rate than variants with six changes. A descendant of BQ.1 called BQ.1.1 has six such changes, and is rising rapidly across Europe, North America and other places.
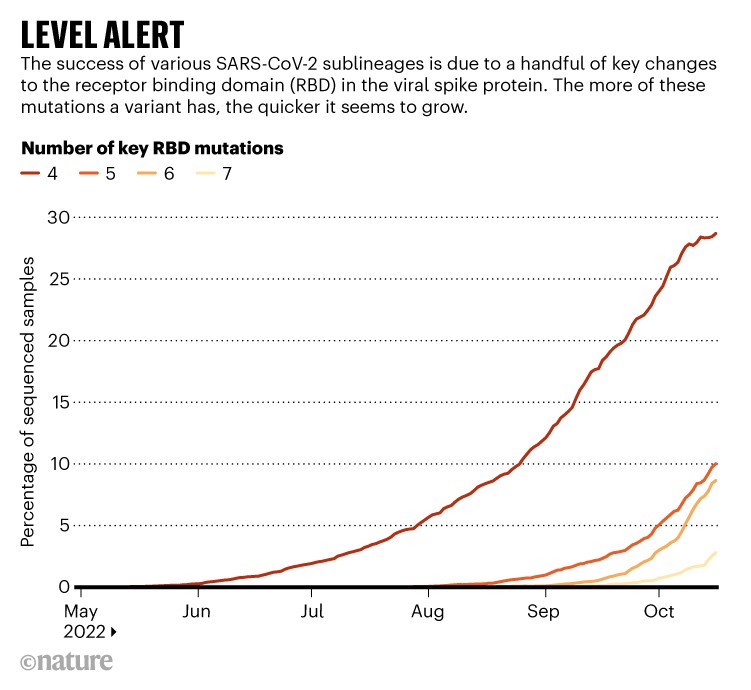
A seventh RBD change seems to lead to still swifter growth (although scientists caution that the estimates are approximate, particularly when the number of sequences recorded is small). The main ‘level 7’ variant scientists are tracking is XBB. The sub-lineage is a hybrid, or recombinant, of two Omicron sublineages, both descendants of BA.2.
Of the swarm, BQ.1.1 and XBB seem to be rising to the top. The BQ.1 family is already dominant in France and is likely to drive infection waves across Europe and North America as these regions enter winter. It is also a common ingredient of the variant soup in South Africa, Nigeria and elsewhere in Africa. XBB, by contrast, looks likely to dominate infections in Asia, where it recently drove a wave of infections in Singapore.
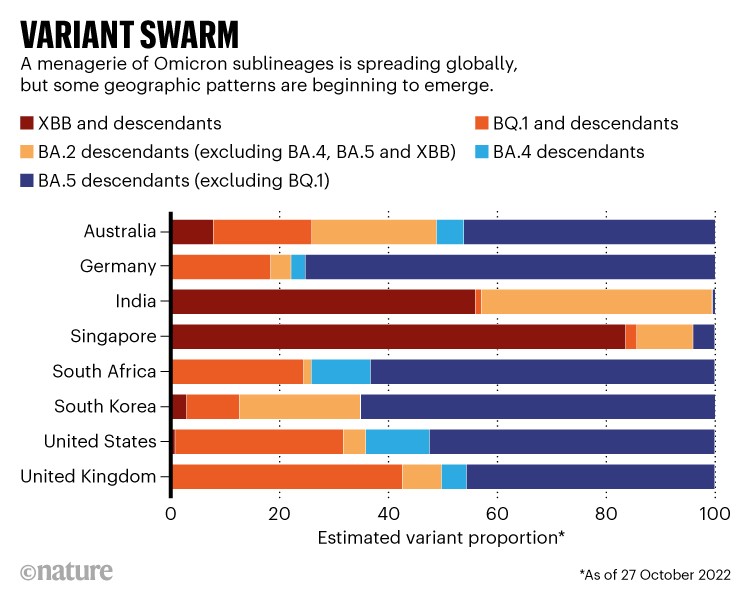
Researchers are also monitoring countries where BQ.1.1 and XBB are co-circulating, to see which spreads faster. In Australia, there are some early signs that XBB is gaining an edge, notes Roemer. This also seems to be happening in India, according to Rajesh Karyakarte, a microbiologist based at the BJ Government Medical College in Pune, who coordinates SARS-CoV-2 genetic sequencing in the state of Maharashtra. “We will be in a position to tell which one survives here. We suspect XBB.”

What Omicron’s BA.4 and BA.5 variants mean for the pandemic
XBB’s advantage over the BQ.1 family might be due in part to changes outside the spike RBD, says Cao. The variant also has mutations in part of the genome encoding a region of the spike protein called the N-terminal domain (NTD). Our immune systems also target this portion of spike with neutralizing antibodies, and people who have recovered from BA.2 and BA.5 infections mount especially strong immune responses to NTD, according to preliminary data from Cao’s lab.
XBB’s ability to dodge antibodies targeting the NTD might allow it to infect people who were immune to BQ.1 and its relatives, Cao adds. But “BQ.1 is picking up NTD mutations crazily fast”, he says. Unpublished work from his team suggests that such additions substantially enhance those variant’s ability to evade neutralizing antibodies raised by vaccination and previous infection.
It’s possible that BQ.1.1 will cause a spike in cases, only for XBB to overtake it in some places, says Roemer. “If it turns out that XBB is going to dominate globally in the end, we might see some sort of double wave in Europe and North America,” he says.
Double immunity?
A key determinant will be the extent to which infection with BQ.1 lineages protect against XBB. Cao’s team is currently working on this. “I have a feeling that if you’re infected with BQ.1, you might have some protection against XBB,” he says. “We don’t have data yet.”
Whether driven by XBB, BQ.1.1 or another member of the swarm, large infection waves can disrupt society, and even mild infections might result in long-lasting health effects. But researchers are keeping an especially close eye on whether the coming waves lead to high numbers of hospitalizations and deaths.
The quest to find genes that drive severe COVID
In an unpublished, preliminary study of 28 people with XBB infections, Karyakarte’s team found that none had severe symptoms. Karyakarte says his colleagues in Bangladesh report similar patterns. Singapore has recorded a small rise in COVID-19 hospitalizations and deaths during its XBB wave, but these severe effects have been smaller than in past waves.
But factors such as seasonality — the Northern Hemisphere winter weather is likely to give SARS-CoV-2 circulation a boost — prior waves, and policy mean that Singapore’s experience might not predict what other countries are in for, says Roemer. “It’s probably not a blueprint for what’s going to happen.”
The Link LonkOctober 29, 2022 at 01:10AM
https://news.google.com/__i/rss/rd/articles/CBMiMmh0dHBzOi8vd3d3Lm5hdHVyZS5jb20vYXJ0aWNsZXMvZDQxNTg2LTAyMi0wMzQ0NS020gEA?oc=5
COVID 'variant soup' is making winter surges hard to predict - Nature.com
https://news.google.com/search?q=hard&hl=en-US&gl=US&ceid=US:en
No comments:
Post a Comment